
본 포스팅은 <파이썬 머신러닝 완벽 가이드>의 3장 내용을 참고하였습니다.
전체적인 데이터를 살피고, 로지스틱 회귀 모형 결과를 교정하는 내용을 담고 있습니다.
1. 데이터 불러오고, 결괏값 분포 체크
from sklearn.linear_model import LogisticRegression
import pandas as pd
diabetes_data = pd.read_csv('diabetes.csv')
print(diabetes_data['Outcome'].value_counts())
diabetes_data.head(3)
diabetes_data.info()
2. 결과 평가 함수 정의
- 정확도, 정밀도, 재현율, f1 값, roc_auc 값, 혼동행렬 출력
- 입력 파라미터는 y_test, 예측 결과(pred), 1로 예측할 확률(pred_proba)
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score, confusion_matrix, roc_auc_score
def get_clf_eval(y_test, pred=None, pred_proba=None):
confusion = confusion_matrix( y_test, pred)
accuracy = accuracy_score(y_test , pred)
precision = precision_score(y_test , pred)
recall = recall_score(y_test , pred)
f1 = f1_score(y_test,pred)
# ROC-AUC 추가
roc_auc = roc_auc_score(y_test, pred_proba)
print('오차 행렬')
print(confusion)
# ROC-AUC print 추가
print('정확도: {0:.4f}, 정밀도: {1:.4f}, 재현율: {2:.4f},\
F1: {3:.4f}, AUC:{4:.4f}'.format(accuracy, precision, recall, f1, roc_auc))
3. 1차 모델링(로지스틱 회귀)
- 피처, 레이블 데이터 세트 분리
- train_test_split 으로 학습/평가 데이터 분리
- 로지스틱 회귀로 학습, 예측, 평가 ➡ 예측값과, 클래스 1에 대한 예측 확률값 도출
- 2에서 정의한 평가 함수로 결과 확인
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
# 피처 데이터 세트(x), 레이블 데이터 세트(y)를 추출
X = diabetes_data.iloc[:, :-1]
y = diabetes_data.iloc[:, -1]
# 학습/평가 데이터 분리(불균형 데이터 -> 층화추출)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=156, stratify = y)
# 로지스틱 회귀로 학습, 예측, 평가
lr_clf = LogisticRegression(solver='liblinear')
lr_clf.fit(X_train, y_train)
pred = lr_clf.predict(X_test)
pred_proba = lr_clf.predict_proba(X_test)[:, 1]
get_clf_eval(y_test, pred, pred_proba)
4. 임곗값 변화에 따른 재현율과 정밀도 시각화(함수)
- precision_recall_curve 사용
- 시각화 함수 정의 : 매개변수는 y_test, 클래스 1값으로의 예측 확률
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
from sklearn.metrics import precision_recall_curve
import numpy as np
def precision_recall_curve_plot(y_test, pred_proba_c1):
# threshold ndarray와 이 threshold에 따른 정밀도, 재현율 ndarray 추출.
precisions, recalls, thresholds = precision_recall_curve(y_test, pred_proba_c1)
plt.figure(figsize = (8, 6))
threshold_boundary = thresholds.shape[0]
plt.plot(thresholds, precisions[0:threshold_boundary], linestyle='--', label = 'precision')
plt.plot(thresholds, recalls[0:threshold_boundary], label = 'recall')
start, end = plt.xlim()
plt.xticks(np.round(np.arange(start, end, 0.1), 2))
plt.xlabel('threshold value')
plt.ylabel('Precision and Recall Value')
plt.legend()
plt.grid()
plt.show()- 함수 적용
pred_proba_c1 = lr_clf.predict_proba(X_test)[:, 1]
precision_recall_curve_plot(y_test, pred_proba_c1)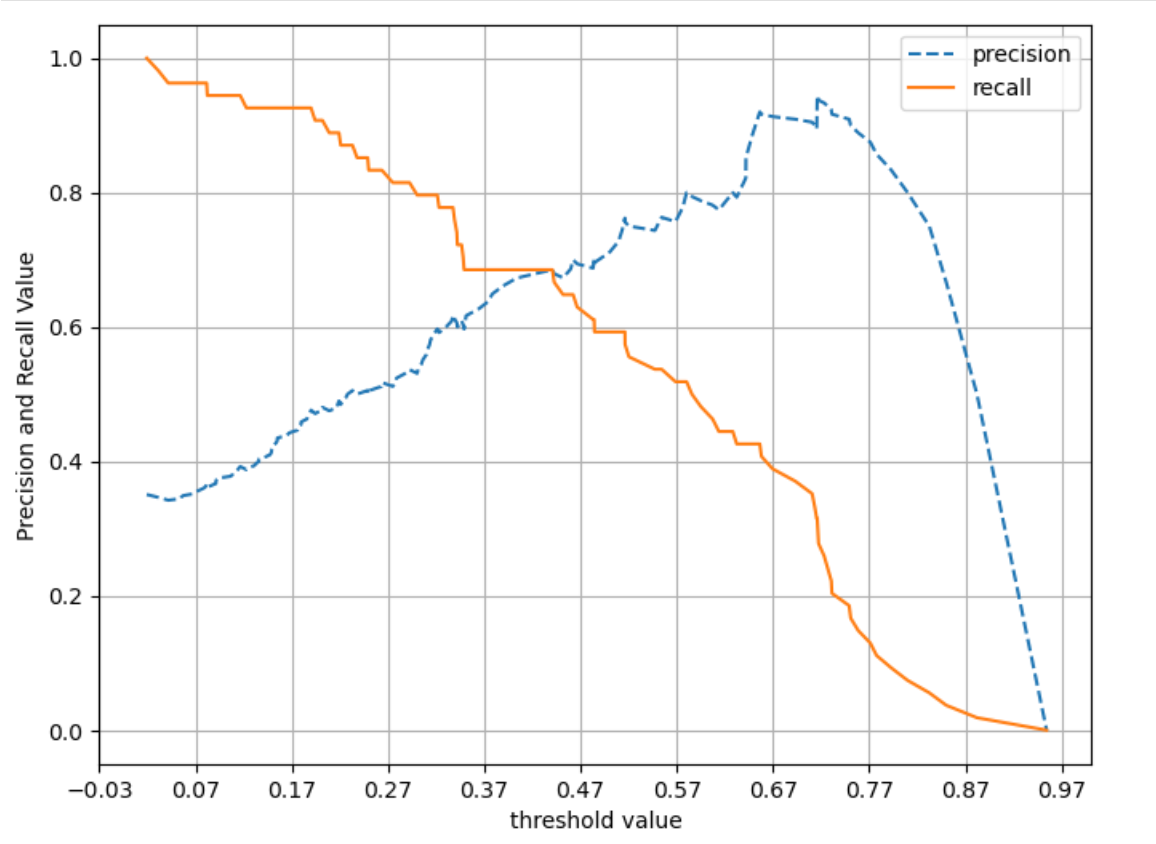
두 그래프가 만나는 지점의 정밀도와 재현율이 모두 0.8 이하로 낮은 편이므로, 다시 한번 전체 데이터 검토
5. 데이터 분포 재확인
diabetes_data.describe()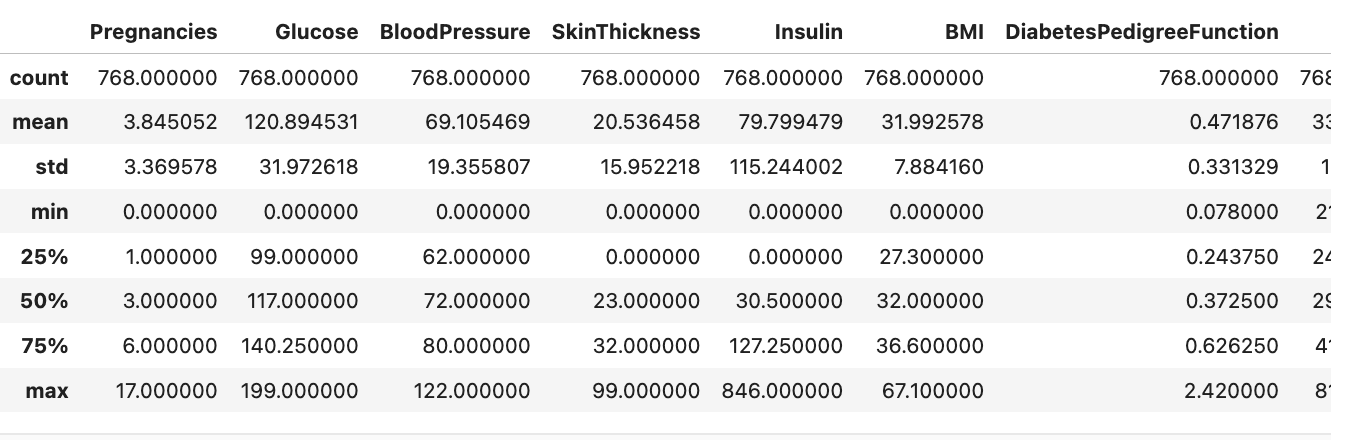
👉 일부 컬럼의 최솟값이 0인 것을 확인 : 0인 것이 불가능한 컬럼의 이상치로 판단
👉 히스토그램을 그려서 좀 더 자세히 파악해보자.(Glucose 컬럼)
plt.hist(diabetes_data['Glucose'], bins = 100)
plt.show()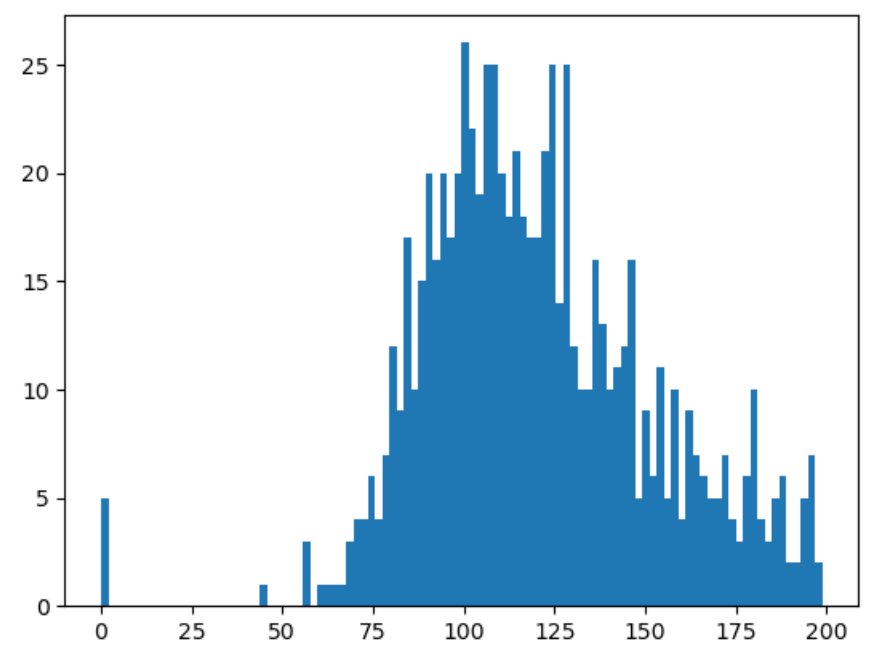
👉 확실히 0값이 보임
👉 0 값을 처리할 필요가 있음 ->> 평균으로 대체
6. 0값이 있는 컬럼의 0값 비율 조사(전체 데이터 대비)
- 0값이 있을 수 없는 피처명을 리스트로 정리
- 전체 데이터 건수 측정
- 0값 검사할 피처들 각각의 총 0값의 개수를 집계
- 전체 값 대비 0값의 비율을 출력
# 0값을 검사할 피처명
zero_features = ['Glucose', 'BloodPressure', 'SkinThickness', 'Insulin', 'BMI']
# 전체 데이터 건수
total_count = diabetes_data['Glucose'].count()
# 피처별로 반복하며, 데이터 값이 0인 데이터 건수 추출, 비율 계산
for feature in zero_features:
zero_count = diabetes_data[diabetes_data[feature] == 0][feature].count()
print('{0}피처의 0 건수는 {1}, 퍼센트는 {2:.2f}%'.format(feature, zero_count, 100*zero_count/total_count))
7. 0값이 있는 컬럼을 평균으로 대체
- 전체 컬럼 중 0값 대체가 필요한 컬럼(zero_features)의 평균을 도출
- replace() 함수로 zero_features의 0값을 평균값으로 대체
# 0값을 평균값으로 대체!
mean_zero_features = diabetes_data[zero_features].mean()
diabetes_data[zero_features] = diabetes_data[zero_features].replace(0, mean_zero_features)8. StandardScaler 후, 로지스틱 회귀 재시도
- ✋ 로지스틱회귀는 일반적으로 숫자 데이터에 스케일링을 적용하는 것이 좋음
- 다시 한 번, X, y값을 나누고 X 값에 대해 StandardScaler wjrdyd
- train, test 데이터셋 분리
- 로지스틱 회귀로 학습, 예측, 평가(평가는 위에서 만든 함수: get_clf_eval 사용)
from sklearn.preprocessing import StandardScaler
X = diabetes_data.iloc[:, :-1]
y = diabetes_data.iloc[:, -1]
# StandardScaler 클래스로 피처 데이터셋에 일괄 스케일링
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(X_scaled, y, test_size = 0.2, random_state=156, stratify=y)
#로지스틱 회귀로 학습, 예측, 평가
lr_clf = LogisticRegression()
lr_clf.fit(X_train, y_train)
pred = lr_clf.predict(X_test)
pred_proba = lr_clf.predict_proba(X_test)[:, 1]
get_clf_eval(y_test, pred, pred_proba)
9. 재현율이 낮은 문제 해결 --- 임곗값을 변화시키며 결과 확인
- Binarizer 메서드로 threshold(임곗값)을 변화시키며 predict 결괏값을 다르게 하여
- 평가 결과를 확인하는 함수 작성
from sklearn.preprocessing import Binarizer
def get_eval_by_thresholds(y_test, pred_proba_c1, thresholds):
for custom_threshold in thresholds:
binarizer = Binarizer(threshold = custom_threshold).fit(pred_proba_c1)
custom_predict = binarizer.transform(pred_proba_c1)
print('임곗값: ', custom_threshold)
get_clf_eval(y_test, custom_predict, pred_proba_c1)
print('\n')- 함수 적용
thresholds = [0.3, 0.33, 0.36, 0.39, 0.42, 0.45, 0.48, 0.50]
pred_proba = lr_clf.predict_proba(X_test)
get_eval_by_thresholds(y_test, pred_proba[:, 1].reshape(-1, 1), thresholds)
10. 정밀도와 재현율이 적절히 높은 지점 확인(임곗값 0.48)
-->> 임곗값 0.48로 최종 로지스틱 회귀 진행 🚀
# 임곗값을 0.48로 설정한 Binarizer 생성
binarizer = Binarizer(threshold=0.48)
pred_th_048 = binarizer.fit_transform(pred_proba[:, 1].reshape(-1, 1))
get_clf_eval(y_test, pred_th_048, pred_proba[:, 1])
728x90
'Machine Learning > Case Study 👩🏻💻' 카테고리의 다른 글
| [Kaggle] 이커머스 데이터 분석 1 (CRM Analytics 🛍️🛒) (0) | 2023.10.08 |
|---|---|
| [공모전 수상작 스터디] 일회용품 쓰레기 감소를 위한,다회용기 비즈니스 모델 개발 (서울시 빅데이터 캠퍼스) (1) | 2023.10.05 |
| [🦀 게 나이 예측(6)] Baseline Modeling 2 (LAD Regression) (0) | 2023.09.26 |
| [🦀 게 나이 예측(5)] 모델 성능 개선을 위한 Feature Engineering (2) | 2023.09.25 |
| [🦀 게 나이 예측(4)] Baseline Modeling(여러 모델 결과를 종합, 비교) (0) | 2023.09.25 |



